Bio-JOIE: Joint Representation Learning of Biological Knowledge Bases
Abstract
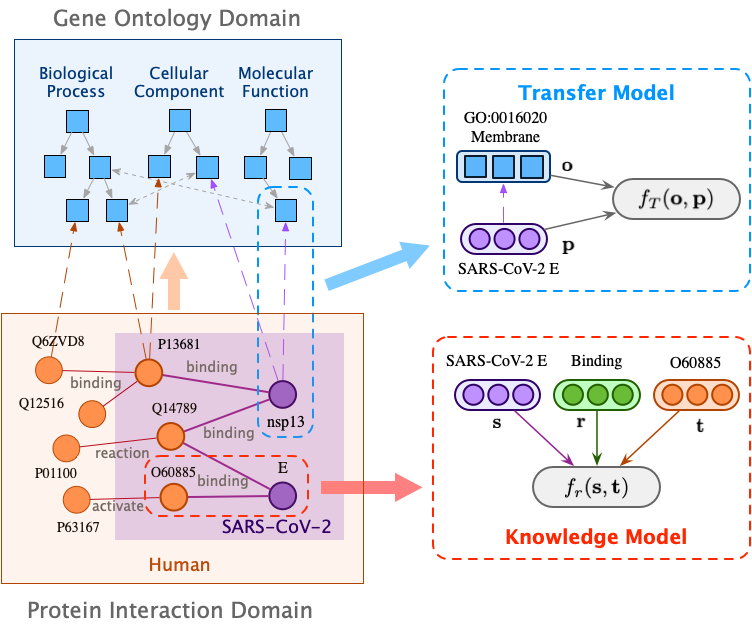
The widespread of coronavirus has led to a worldwide pandemic with a high mortality rate. Currently, the knowledge accumulated from different studies about this virus is very limited. Leveraging a wide-range of biological knowledge, such as gene ontology and protein-protein interaction (PPI) networks from other closely related species presents a vital approach to infer the molecular impact of a new species. In this paper, we propose the transferred multi-relational embedding model Bio-JOIE to capture the knowledge of gene ontology and PPI networks, which demonstrates superb capability in modeling the SARS-CoV-2-human protein interactions. Bio-JOIE jointly trains two model components. The knowledge model encodes the relational facts from the protein and GO domains into separated embedding spaces, using a hierarchy-aware encoding technique employed for the GO terms. On top of that, the transfer model learns a non-linear transformation to transfer the knowledge of PPIs and gene ontology annotations across their embedding spaces. By leveraging only structured knowledge, Bio-JOIE significantly outperforms existing state-of-the-art methods in PPI type prediction on multiple species. Furthermore, we also demonstrate the potential of leveraging the learned representations on clustering proteins with enzymatic function into enzyme commission families. Finally, we show that Bio-JOIE can accurately identify PPIs between the SARS-CoV-2 proteins and human proteins, providing valuable insights for advancing research on this new disease.
Modeling
More Info
Publication: The 11th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (BCB 2020)
Date: September, 2020
Links: ACM Library, PDF, Video, Slides. Dataset and code are available here.